Diversity of microbial communities
The number of microbial cells in the oceans is estimated to be 1029 and in the whole Planet there may be 1030 cells. However, we do not know to how many different species do they belong. The ecologist Robert May stated “we have a catalog of all the objects that our instruments can detect in the Universe, but we ignore how many living beings share the Earth with us.” In effect, estimates of bacterial species in the oceans, for example, range from a few thousand to 1010.
We are interested in two particular aspects of microbial diversity. The first one is to understand the distribution and abundances of the different species in a community. This distribution can be illustrated with a rank-abundance curve. Obviously, a few species are very abundant and likely contribute to carbon and energy flow in the ecosystem. But most species are rare and in some cases extremely rare. By being rare, these taxa are likely protected from viral lysis and predator attacks and can, thus, persist for long periods of time. This confers to microbial communities their particular properties of resilience and adaptability. We are studying microbial diversity in marine samples as well as in extreme environments.
The second point is to unravel the structure of taxa. Usually, operational taxonomic units (OTUs) are considered in ecological studies. The definition of an OTUs was traditionally those sequences of 16S rDNA that were at least 97% similar. However, this threshold is now considered too low, since a lot of significant diversity can be found even at a 99% similarity. We study this micro diversity in microbial communities with different approaches. Some methods look at single base differences and estimate the likelihood that they represent real differences and not sequencing errors. These methods use 16S rDNA sequences either from amplicons or from miTAGs.
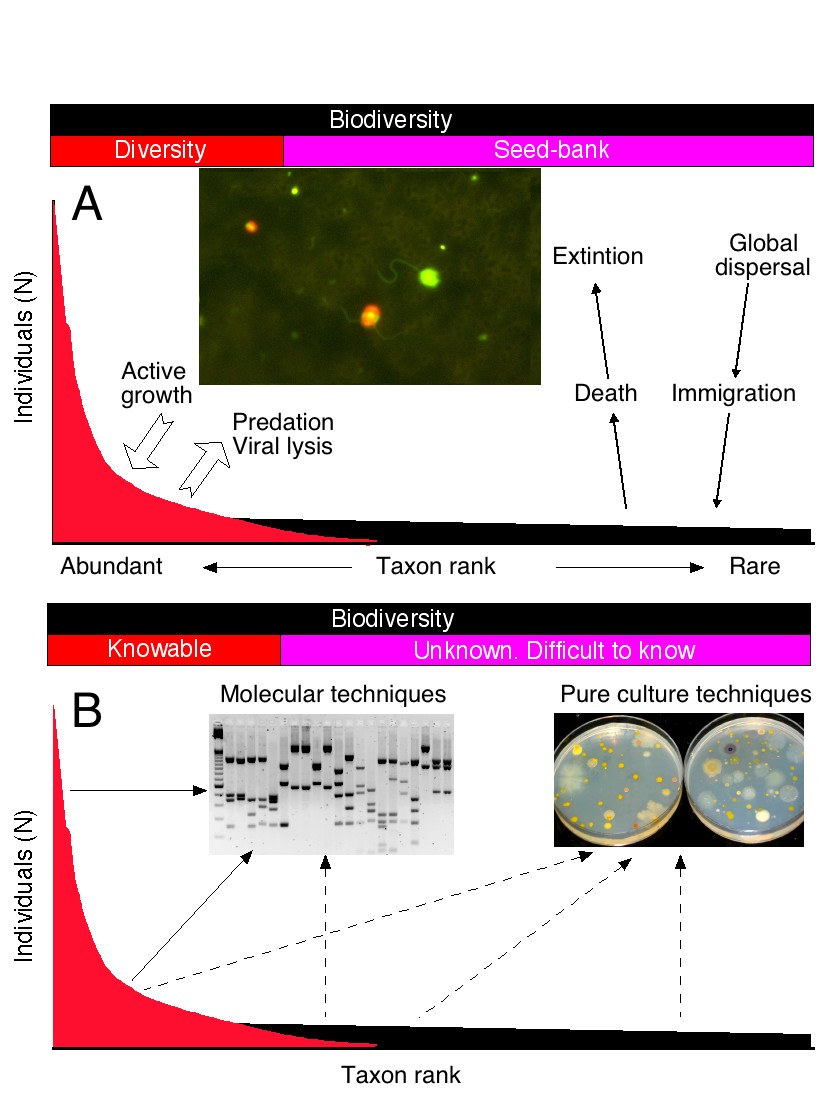
Rank-abundance curves showing the role of different taxa in the ecosystem (above) and the methods applied to retrieve them
Other methods look at the relative abundances of genes or COGs in different metagenomes and reconstruct the genomes of different strains. In this case, annotated metagenomes are used as primary data and methods such as metagenotyping or metagenomic deconvolution are used to retrieve the genomes of different strains. We carry out these studies in freshwater, marine, and Polar microbial communities
Assembly of microbial communities: interactions among microbial taxa
Communities are composed of microbial taxa that necessarily interact among themselves, either in positive or negative ways. Understanding such interactions will eventually help in manipulating communities for desired purposes. For example, in bioremediation, in biotechnology, in human health and even in terraforming. We use three different approaches to identify interacting microorganisms.
One approach consists of building co-occurrence networks. The assumption is that taxa that appear together a number of times significantly above what could be expected by chance are likely partners of an interaction. We use an approach that sequentially builds modules of two to 10 or so taxa that co-occur and then we look for their genomic potential to see whether they can complement each other or compete with each other. We have used the extensive information in our data base EnvDB (link) to build such networks for many different environments, including human gut, animal associated microbiotas, aquatic or terrestrial samples. Many of the environments are very rich comprising thousands of species.
A second approach also uses co-occurrences of microbial taxa. However, in this case, the abundance of the different taxa is followed through time. Microbes that oscillate in abundance in concert, are likely to interact with each other. This information can be extracted with generalized Lotka-Volterra equations. We use this approach in an extreme environment: the crystallizer ponds in solar salterns. The microbial community in these environments is extremely simple, including only 10 to 20 species and, thus, making the analysis possible.
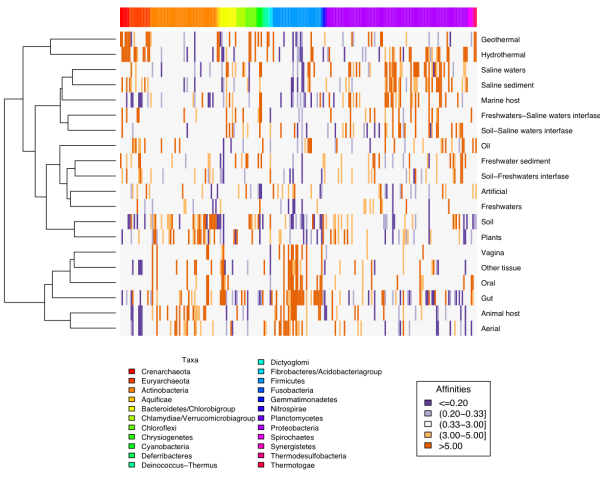
Co-occurrence network of bacteria and fungui in human milk
Finally, we assume that bacteria that are in close physical proximity will also be candidates to be interacting. In this case, we dilute a natural community and encapsulate aliquots containing about 10 to 100 cells in microspheres. These microspheres can be manipulated by flow cytometry, they can be incubated under different conditions, and the bacteria growing together can be sequenced and their metabolisms analyzed. We do these studies with gut microbiome that comprises an intermediate level of complexity with a few hundred species present.
In all these cases, the candidate members of an interacting group can be sequenced and their genomes used in metabolic models to analyze their potential interactions.
Functioning of microbial communities
Genomes of environmentally relevant bacteria.
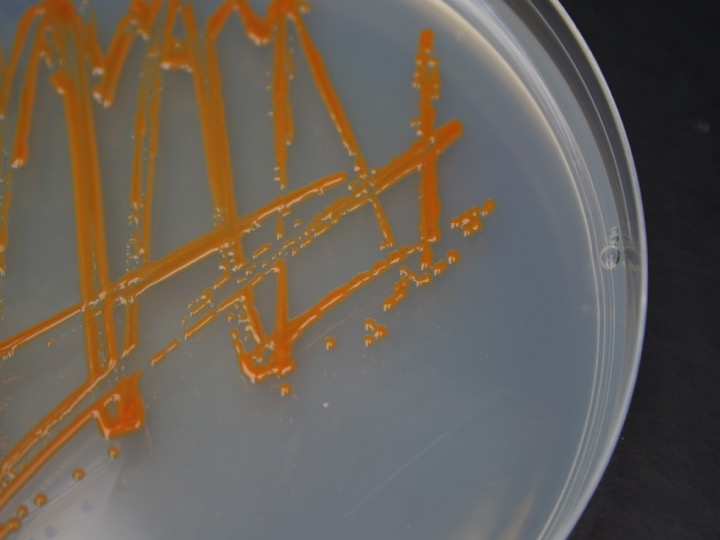
There are three ways to retrieve the genomes of environmentally relevant microorganisms. The first is to obtain the microorganism in pure culture such as those of Dokdonia MED132 and Polaribacter MED152 shown in the figure. This allows physiological experiments as well as sequencing of the complete genomes. We have worked on marine Flavobacteria.
Unfortunately, many abundant bacteria are difficult to isolate. There are two alternatives to culturing. In one approach, single cells are sorted into individual wells by flow cytometry. The DNA can then be amplified and sequenced, obtaining a SAG (single amplified genome). We are using this approach in Polar waters looking for marine Flavobacteria.
Finally, a third option is to assemble a genome from a metagenome obtaining a MAG. In this case, the reads of a metagenome are assembled into contigs and these are binned into taxonomic categories. In many cases almost complete genomes can be recovered. We are doing these studies with samples from both hot springs and Polar waters.
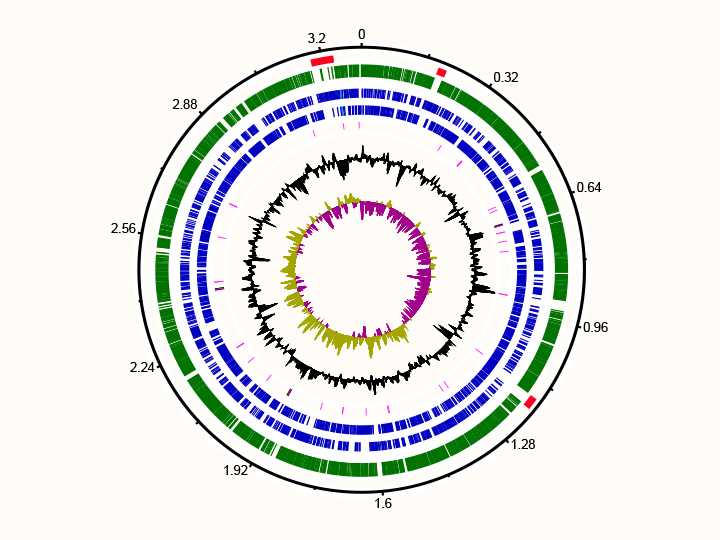
Two marine Flavobacteria with proterhodopsins: pure culture of Polaribacter MED132 (left) and genome of Dokdonia MED134 (right)
Metagenomes and metatranscriptomes
In principle, metagenomes include all the genes from all the microorganisms in a sample or, at least, those of the abundant organisms. The short reads obtained after sequencing can be assembled and annotated. Then, different kinds of analyses are possible. The taxa responsible for a given function, for example nitrogen fixation, can be identified. Or the abundance of genes for a particular function such as photosynthesis can be followed with time. We are using metagenomics and metatranscriptomics to reconstruct the functioning of microbial mats in hot springs, to analyze the relevant genes being expressed in Polar waters from winter to summer, or to study the genomic organization of proteorhodopsin genes in marine bacteria.